|
 |
- Search
| Asian Spine J > Volume 10(5); 2016 > Article |
Abstract
Several studies have evaluated the association between vitamin D receptor, matrix metalloproteinase 3 (MMP-3) polymorphisms and the risk of intervertebral disc degeneration susceptibility. The findings were inconsistent. This meta-analysis aimed to systematically assess the association between vitamin D receptor, MMP-3 polymorphisms and the risk of intervertebral disc degeneration susceptibility. A search of various databases was done covering all papers published until December 31th, 2014. Eight, 4, 3 studies were finally included that addressed the risk of intervertebral disc degeneration susceptibility and vitamin D receptor FokI (rs2228570), ApaI (rs7975232), and MMP-3 (rs731236) polymorphisms, respectively. FokI (f vs. F: summary odds ratio [OR], 1.13; 95% confidence interval [CI], 0.76ŌĆō1.69; ff vs. FF: OR, 1.02; 95% CI, 0.59ŌĆō1.77; ff vs. Ff/FF: OR, 1.05; 95% CI, 0.70ŌĆō1.58), ApaI (a vs. A: OR, 0.73; 95% CI, 0.45ŌĆō1.19; aa vs. AA: OR, 0.53; 95% CI, 0.22ŌĆō1.25 p=0.14; aa vs. AA/Aa: OR, 0.69; 95% CI, 0.53ŌĆō0.89) in the vitamin D receptor gene and MMP3 polymorphisms (5A vs. 6A: OR, 1.92; 95% CI, 0.77ŌĆō4.80; 5A5A vs. 6A6A: OR, 2.17; 95% CI, 0.75ŌĆō6.24; 5A5A vs. 5A6A/6A6A: OR, 1.58; 95% CI, 0.72ŌĆō3.44) were not obviously associated with risk of intervertebral disc degeneration susceptibility. FokI, ApaI polymorphisms in the vitamin D receptor gene and MMP-3 polymorphism are not obvious risk factors for intervertebral disc degeneration susceptibility.
Low back pain (LBP) is universally seen in orthopedic practices. It is one of the most influential factors affecting quality of life and exacts a significant social and economic impact worldwide. It is estimated that 50%ŌĆō80% adults will experience at least one episode of back pain during their lifetime. The etiology of LBP is still not fully understood, although intervertebral disc degeneration (IDD) is believed to be a major cause [12].
IDD has both genetic and environmental origins. Environmental factors include body weight, mechanical loading, physical activities and smoking may also have a role in the development of IDD. Epidemiological studies on families and twins have provided evidence that inheritance is the major determinant of IDD susceptibility [34]. Genes associated with degenerative disc disease in human include genes coding for vitamin D receptor (VDR); matrix metalloproteinase 3 (MMP-3); collagens I, IX, and XI; interleukin-1; aggrecan and other proteins [5].
VDR is an important factor in bone metabolism and development. Vitamin D facilitates the intestinal absorption of calcium through the activated VDR, stimulates renal production of 1, 25-(OH)2-vitamin D3 and influences osteoblasts, osteoclasts, and secretion of parathyroid hormone [4]. The 100 kb VDR gene is located on human chromosome 12 (12q12ŌĆōq14), and has more than 100 restriction endonuclease cutting site polymorphisms [46]. Select polymorphic sites that have been studied to assess the association between VDR gene polymorphism and risk of IDD susceptibility include FokI (rs2228570) and ApaI (rs7975232) restriction sites [4789101112]. However, results have been contradictory conclusions; some studies suggested an association of VDR gene polymorphism with the risk of IDD susceptibility [410], while others did not [8912].
MMP-3 mediates disc degeneration by degrading matrix proteoglycans and collagens. The common MMP-3 gene polymorphism is 5A/6A (rs72520913). The polymorphism occurs in the promoter region of the human MMP-3 gene; one allele has a run of six adenosines (6A) and the other allele has a run of five (5A) [61314]. Several recent studies that assessed the association between MMP-3 (rs731236) polymorphism and the risk of IDD susceptibility provided equivocal findings with an association evident in several studies [141516] but not in another [17].
To provide more clarity, we conducted a meta-analysis.
Searches of PubMed, Web of Science, China National Knowledge Infrastructure (CNKI) and WangFang databases were done without language or publication year limitation. The last search was updated on December 31, 2014. Search terms were "vitamin D receptor" or "VDR" or "matrix metalloproteinase-3" or "MMP-3", and "polymorphisms" and either "intervertebral disc degeneration" or "IDD". The reference lists of review articles, clinical trials, and meta-analysis were also screened manually.
Inclusion criteria for articles were: (1) evaluation of the associations between VDR (FokI or ApaI) polymorphisms or MMP-3 polymorphism and the risk of IDD susceptibility; (2) case-control design; (3) sufficient data for cases and controls and (4) allele distributions met HardyŌĆōWeinberg equilibrium (HWE). The exclusion criteria were: (1) not a case-control study, (2) unavailable genotype frequency and (3) duplication of previous publication. Two investigators carefully and independently reviewed all identified studies to determine whether an individual study was eligible for inclusion in this meta-analysis.
Data were extracted independently by both two investigators (Y.J.H. and S.J.Z.) using a pre-designed form that contained: (1) name of the first author, (2) year of publication, (3) number of genotypes in cases and controls, (4) country of origin and ethnicity of the study population, (5) source of control subjects, (6) numbers of cases and controls and (7) gender and age of enrolled subjects. Any disagreement was resolved by consensus.
The Newcastle-Ottawa Scale (NOS), which is one of the most useful tool for assessing methodological quality of non-randomized studies, was used to assess the risk of bias in the included non-randomized studies. "Selection" (4 items), "comparability" (1 item), "exposure" (3 items) were evaluated, and each "high" quality choice was awarded a "star".
Review Manger 5.0 software was used for data analysis. For each study, the odds ratios (ORs) and 95% confidence intervals (CIs) were used to evaluate the association. Heterogeneity was estimated by the I2 statistic. If the value of I2>50%, the Mantel-Haenszel random-effects model (REM) was used, the source of heterogeneity was investigated by a subgroup analysis and a sensitivity analysis. Alternately, the fixed-effects model (FEM) was used to estimate the pooled OR. Subgroup analysis was performed according to different ethnicity or age. The sensitivity analysis was performed by rejecting each article with high statistical heterogeneity [18]. A p<0.05 was defined as statistically significant.
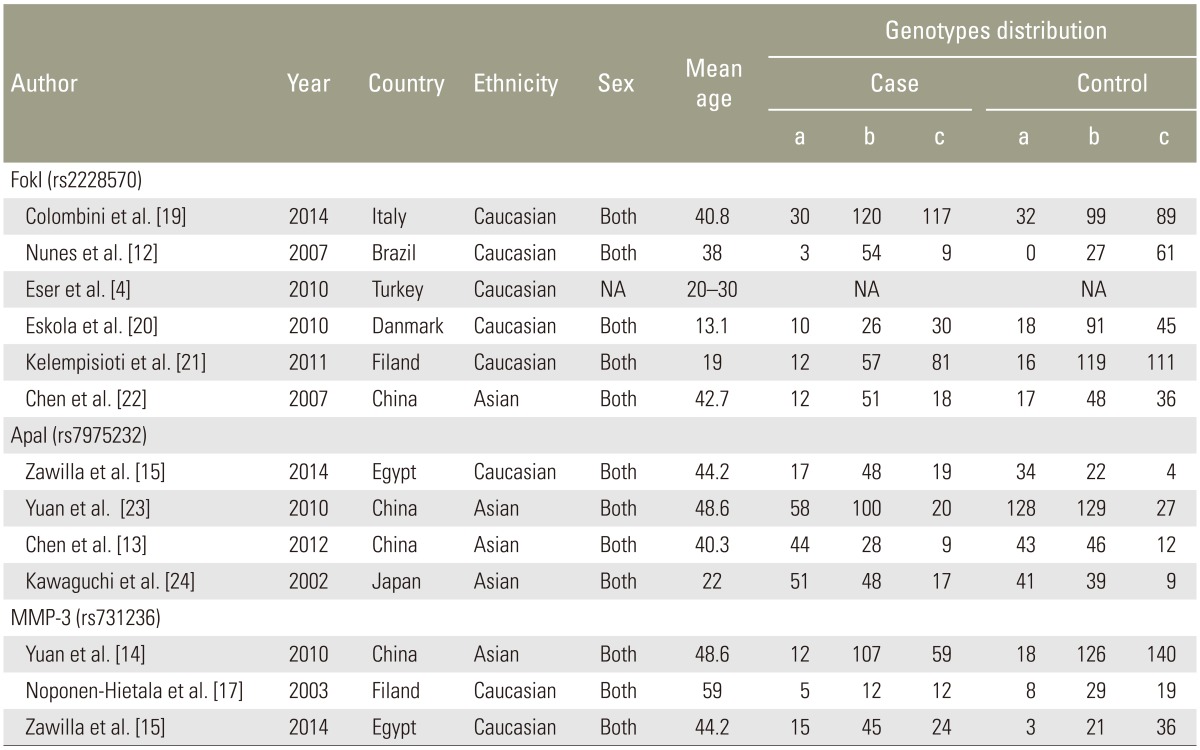
Fig. 1 presents the search results. Fifteen studies were identified involving the association between the risk of IDD susceptibility and VDR FokI (n=8), ApaI (n=4), and MMP-3 (n=3) polymorphisms. Asian populations were assessed concerning VDR FokI polymorphism in 2 studies and ApaI polymorphism in one study. The remaining studies recruited Caucasians. For the 3 studies that assessed MMP-3 polymorphism, 2 studies evaluated Caucasians and Asians. All subjects included in the studies had blood drawn for DNA extraction. One study used computed tomography (CT) for the detection of IDD [15], with magnetic resonance imaging (MRI) used in other studies. VDR FokI and ApaI polymorphism genotype distribution in control groups were in HWE [18]. The detailed characteristics of the included studies are summarized in Table 1.
The results of the study types and NOS assessments are provided in Table 2. All of the studies were case-control studies. A range of 5ŌĆō7 stars was gained by NOS, indicating median to high methodological quality of the included studies.
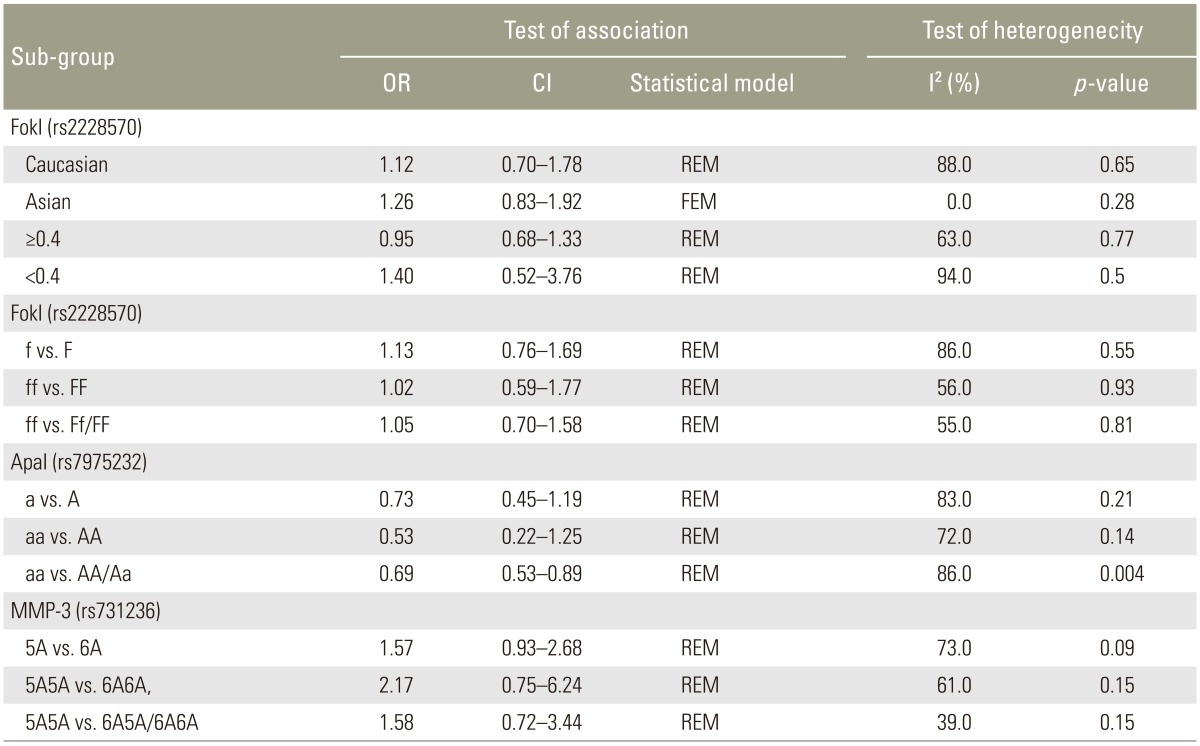
Eleven studies reported the association between the VDR (FokI, ApaI) polymorphisms and the risk of IDD susceptibility. Characteristics of the individual studies are summarized in Table 1. In the pooled analysis for the association between VDR FokI polymorphism and the risk of IDD susceptibility, the I2 was 86%; the significant heterogeneity was assessed and found not to be a concern, so a randomeffect approach was used to estimate ORs and CIs. No significant association between VDR FokI polymorphism and the risk of IDD susceptibility was evident (Table 3) (f vs. F: OR, 1.13; 95% CI, 0.76ŌĆō1.69; p=0.55; ff vs. FF: OR, 1.02; 95% CI, 0.59ŌĆō1.77, p=0.93; ff vs. Ff/FF: OR, 1.05; 95% CI, 0.70ŌĆō1.58; p=0.81). Subgroup analysis stratified by ethnicity and age also confirmed the above results, with no significant association evident in Caucasians (f vs. F: OR, 1.12; 95% CI, 0.70ŌĆō1.78, p=0.64) or Asians (f vs. F: OR, 1.26; 95% CI, 0.83ŌĆō1.92). No associations were evident with age Ōēź40 years (f vs. F: OR, 0.95; 95% CI, 0.68ŌĆō1.33; p=0.77) or age <40 years (f vs. F: OR, 1.40; 95% CI, 0.52ŌĆō3.76; p=0.50). Sensitivity analysis was performed by the sequential removal of the studies; no one study influenced the results significantly.
ORs and CIs results were also estimated using a random-effect approach owing to the significant heterogeneity (I2=83%). The pooled results indicated no association between VDR Apal polymorphism and the risk of IDD susceptibility too (Table 3) (a vs. A: OR, 0.73; 95% CI, 0.45ŌĆō1.19, p=0.21; aa vs. AA: OR, 0.53; 95% CI, 0.22ŌĆō1.25, p=0.14; aa vs. AA/Aa: OR, 0.69; 95% CI, 0.53ŌĆō0.89, p=0.04). The sensitivity analysis confirmed the above conclusion. As limited studies were included for the investigation, subgroup analysis could not be performed.
Results of the association between MMP-3 polymorphism and the risk of IDD susceptibility are presented in Table 2. No significant association was evident (5A vs. 6A: OR, 1.57; 95% CI, 0.93ŌĆō2.68, p=0.09; 5A5A vs. 6A6A: OR, 2.17; 95% CI, 0.75ŌĆō6.24; p=0.15; 5A5A vs. 5A6A/6A6A: OR, 1.58; 95% CI, 0.72ŌĆō3.44; p=0.15). Sensitivity analysis suggested that no individual study significantly affected the overall OR. We did not perform subgroup analysis because the sample size was too small.
IDD is a multifactorial and chronic process that alters the structure and function of intervertebral discs, which is a common multifactorial and multigenic disease [2023]. The association between gene polymorphism and the etiology of IDD has become a hot research field in recent years. This will bring positive significance for the prevention and treatment of IDD. Some studies have reported at least 14 genes including the VDR and MMP-3 gene that may have more important influence on the susceptibility of IDD [222425]. The association between VDR gene polymorphism and IDD susceptibility has been validated in large populations of diverse ethnic backgrounds. The association between MMP-3 polymorphism and IDD susceptibility has been a hot topic in recent years, with no consensus reached. To provide clarity, we undertook the present meta-analysis.
VDR is a steroid nuclear receptor, which participates in normal bone mineralization and remodeling. The association between VDR polymorphisms and the risk of IDD susceptibility has been investigated for long time. FokI (rs2228570) and ApaI (rs7975232) are two polymorphic sites of VDR gene involved the risk of IDD susceptibility. To our knowledge, only one meta-analysis has been performed, with no significant association evident between VDR polymorphism and the risk of IDD susceptibility [26]. We reached the same conclusion (Figs. 2, 3). Subgroup analysis stratified by ethnicity and age also confirmed the result. Most of the included studies did not eliminate non-genetic risk factors including obesity, smoking and heavy physical occupations, so it remains possible that a potential association was obscured by some environmental factors. Moreover, VDR polymorphisms have been reported to be significantly associated with the multilevel and severe forms of IDD [1327]. Therefore, a conclusion still cannot be made and a large-scale and long-term study is still needed.
MMP-3 is a key enzyme that directly degrades components of the extracellular matrix including proteoglycans, laminin, fibronectin, gelatins and collagens. MMP-3 also indirectly affects degradation of the extracellular matrix by activating other latent MMPs [1928]. This is the first meta-analysis addressing the association between MMP-3 gene polymorphism and the risk of IDD susceptibility. The collective results (Fig. 4) indicate no significant association. Sensitivity analysis revealed that no single study significantly influence the pooled effect. The association between MMP-3 gene polymorphism and the susceptibility of IDD could not be easily excluded since only 3 related studies were included in this meta-analysis; further studies were warranted for a reliable result in the future.
The meta-analysis has been recognized as an effective method to resolve a wide variety of clinical questions by summarizing and reviewing the previously published quantitative research. Still, there are several potential limitations of current meta-analysis. Firstly, this meta-analysis was based on unadjusted effect estimates, so a more precise analysis should be conducted if individual data were available. Secondly, IDD is a multifactor disease determined by the synthetic effect of environment and gene, interactions between genes and between genes and the environment and even between different polymorphic loci of the same gene. These interactions may mask or magnify the function of the involved genes. Thirdly, there are methodological limitations. Although all the included studies were caseŌĆōcontrol designed, not all controls in the studies were age or sex matched, which may have been a cause of the pronounced heterogeneity in this analysis. The age range (teenager to aged may have been another confounding factor. Finally, the different imaging modalities (CT and MRI) used for the detection of IDD may have been confounding.
In summary, the current meta-analysis comprehensively reviewed the association between VDR (FokI, ApaI) and MMP-3 polymorphisms and the risk of IDD susceptibility. None of the polymorphisms were significantly associated with the risk of IDD susceptibility. Since potential confounders could not be ruled out completely, further well-designed large studies particularly referring to gene-gene and gene-environment interactions are warranted.
Notes
Conflict of Interest: No potential conflict of interest relevant to this article was reported.
References
1. Virtanen IM, Karppinen J, Taimela S, et al. Occupational and genetic risk factors associated with intervertebral disc disease. Spine (Phila Pa 1976) 2007 32:1129ŌĆō1134. PMID: 17471097.


2. Rubin DI. Epidemiology and risk factors for spine pain. Neurol Clin 2007 25:353ŌĆō371. PMID: 17445733.


3. Battie MC, Videman T, Gibbons LE, Fisher LD, Manninen H, Gill K. 1995 Volvo Award in clinical sciences. Determinants of lumbar disc degeneration: a study relating lifetime exposures and magnetic resonance imaging findings in identical twins. Spine (Phila Pa 1976) 1995 20:2601ŌĆō2612. PMID: 8747238.


4. Eser B, Cora T, Eser O, et al. Association of the polymorphisms of vitamin D receptor and aggrecan genes with degenerative disc disease. Genet Test Mol Biomarkers 2010 14:313ŌĆō317. PMID: 20367178.


5. Chan D, Song Y, Sham P, Cheung KM. Genetics of disc degeneration. Eur Spine J 2006 15(Suppl 3): S317ŌĆōS325. PMID: 16819621.


6. Cheung KM, Chan D, Karppinen J, et al. Association of the Taq I allele in vitamin D receptor with degenerative disc disease and disc bulge in a Chinese population. Spine (Phila Pa 1976) 2006 31:1143ŌĆō1148. PMID: 16648751.


7. Sambrook PN, MacGregor AJ, Spector TD. Genetic influences on cervical and lumbar disc degeneration: a magnetic resonance imaging study in twins. Arthritis Rheum 1999 42:366ŌĆō372. PMID: 10025932.


8. Kalb S, Martirosyan NL, Kalani MY, Broc GG, Theodore N. Genetics of the degenerated intervertebral disc. World Neurosurg 2012 77:491ŌĆō501. PMID: 22120330.


9. Videman T, Lepp├żvuori J, Kaprio J, et al. Intragenic polymorphisms of the vitamin D receptor gene associated with intervertebral disc degeneration. Spine (Phila Pa 1976) 1998 23:2477ŌĆō2485. PMID: 9854746.


10. Mayer JE, Iatridis JC, Chan D, Qureshi SA, Gottesman O, Hecht AC. Genetic polymorphisms associated with intervertebral disc degeneration. Spine J 2013 13:299ŌĆō317. PMID: 23537453.



11. Uitterlinden AG, Fang Y, Van Meurs JB, Pols HA, Van Leeuwen JP. Genetics and biology of vitamin D receptor polymorphisms. Gene 2004 338:143ŌĆō156. PMID: 15315818.


12. Nunes FT, Conforti-Frose ND, Negrell WF, Souza DR. Genetic and environmental factors involved on intervertebral disc degeneration. Acta Ortop Bras 2007 15:9ŌĆō13.

13. Chen W, Li G, Sun H, et al. Association of vitamin D receptor gene polymorphism in Han people with lumbar degenerative disc disease. Afr J Pharm Pharmacol 2012 6:1211ŌĆō1215.
14. Yuan HY, Tang Y, Liang YX, et al. Matrix metalloproteinase-3 and vitamin d receptor genetic polymorphisms, and their interactions with occupational exposure in lumbar disc degeneration. J Occup Health 2010 52:23ŌĆō30. PMID: 20009418.


15. Zawilla NH, Darweesh H, Mansour N, et al. Matrix metalloproteinase-3, vitamin D receptor gene polymorphisms, and occupational risk factors in lumbar disc degeneration. J Occup Rehabil 2014 24:370ŌĆō381. PMID: 23975061.


16. Tang Y, Yuan HY, Wang ZP, Xu JG, Lei L. The genetic polymorphisms of MMP-3 and VDR on susceptibility of lumbar disc degeneration. Fudan Univ J Med Sci 2007 34:37ŌĆō41.
17. Noponen-Hietala N, Kyll├Čnen E, M├żnnikk├Č M, et al. Sequence variations in the collagen IX and XI genes are associated with degenerative lumbar spinal stenosis. Ann Rheum Dis 2003 62:1208ŌĆō1214. PMID: 14644861.



18. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002 21:1539ŌĆō1558. PMID: 12111919.


19. Colombini A, Brayda-Bruno M, Lombardi G, et al. FokI polymorphism in the vitamin D receptor gene (VDR) and its association with lumbar spine pathologies in the Italian population: a case-control study. PLoS One 2014 9:e97027PMID: 24810167.



20. Eskola PJ, Kjaer P, Sorensen JS, et al. Gender difference in genetic association between IL1A variant and early lumbar disc degeneration: a three-year follow-up. Int J Mol Epidemiol Genet 2012 3:195ŌĆō204. PMID: 23050050.


21. Kelempisioti A, Eskola PJ, Okuloff A, et al. Genetic susceptibility of intervertebral disc degeneration among young Finnish adults. BMC Med Genet 2011 12:153PMID: 22107760.



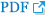
22. Chen WJ, Ye W, Ding Y, et al. Association of vitamin D receptor gene TruI and FokI polymorphisms with lumbar degenerative disc disease in Han nationality orthopedic. J China 2007 15:373ŌĆō375.
23. Yuan HY, Tang Y, Lei L, et al. Synergistic interaction between MMP-3, VDR gene polymorphisms and occupational risk factors on lumbar disc degeneration. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi 2010 28:334ŌĆō338. PMID: 20853681.

24. Kawaguchi Y, Kanamori M, Ishihara H, Ohmori K, Matsui H, Kimura T. The association of lumbar disc disease with vitamin-D receptor gene polymorphism. J Bone Joint Surg Am 2002 84:2022ŌĆō2028. PMID: 12429765.


25. Jones G, White C, Sambrook P, Eisman J. Allelic variation in the vitamin D receptor, lifestyle factors and lumbar spinal degenerative disease. Ann Rheum Dis 1998 57:94ŌĆō99. PMID: 9613338.



26. Xu G, Mei Q, Zhou D, Wu J, Han L. Vitamin D receptor gene and aggrecan gene polymorphisms and the risk of intervertebral disc degeneration:a meta-analysis. PLoS One 2012 7:e50243PMID: 23209686.



27. Balmain N, Hauchecorne M, Pike JW, Cuisinier-Gleizes P, Mathieu H. Distribution and subcellular immunolocalization of 1,25-dihydroxyvitamin D3 receptors in rat epiphyseal cartilage. Cell Mol Biol (Noisy-le-grand) 1993 39:339ŌĆō350. PMID: 8392883.

28. Takahashi M, Haro H, Wakabayashi Y, Kawa-uchi T, Komori H, Shinomiya K. The association of degeneration of the intervertebral disc with 5a/6a polymorphism in the promoter of the human matrix metallo-proteinase-3 gene. J Bone Joint Surg Br 2001 83:491ŌĆō495. PMID: 11380116.


Fig.┬Ā1
Flow chart of study selection based on the inclusion and exclusion criteria. CNKI, China National Knowledge Infrastructure; WOS, Web of Science.
Fig.┬Ā2
Meta-analysis for VDR FokI (rs2228570) polymorphism and the risk of IDD (f vs. F). VDR, vitamin D receptor; IDD, intervertebral disc degeneration; M-H, Mantel-Haenszel; CI, confidence interval.
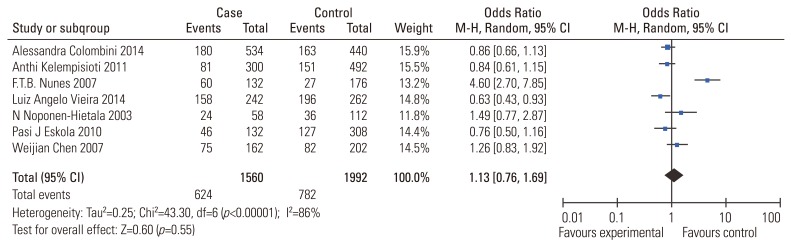
Fig.┬Ā3
Meta-analysis for VDR ApaI (rs7975232) polymorphism and the risk of IDD susceptibility (a vs. A). VDR, vitamin D receptor; IDD, intervertebral disc degeneration; M-H, Mantel-Haenszel; CI, confidence interval.
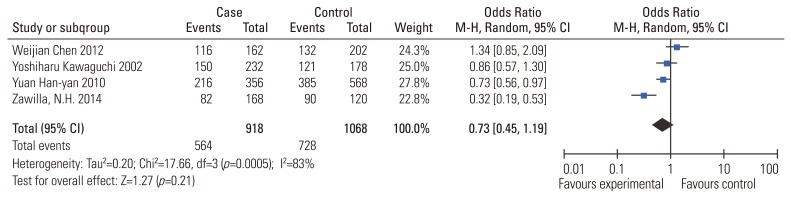
Fig.┬Ā4
Meta-analysis for MMP-3 (rs731236) polymorphism and the risk of IDD susceptibility (5A vs. 6A). MMP-3, matrix metalloproteinase-3; IDD, intervertebral disc degeneration; M-H, Mantel-Haenszel; CI, confidence interval.

Table┬Ā1
Characteristics of individual studies for associations between VDR, MMP-3 polymorphisms and IDD risks
- TOOLS