Novel Biomarkers of Health and Degeneration in Human Intervertebral Discs: In-depth Proteomic Analysis of Collagen Framework of Fetal, Healthy, Scoliotic, Degenerate, and Herniated Discs
Article information
Abstract
Study Design
Profiling proteins expressed in the nucleus pulposus (NP) of intervertebral discs (IVDs) in five different biological states.
Purpose
To evaluate the molecular complexity of the collagen (COL) framework and its role in the health and disease of human IVDs.
Overview of Literature
Changes in COL composition have been linked to degenerative disk disease (DDD). Despite the fact that humans have 28 different types of COLs, most of the literature focuses solely on COL-1 and COL-2. This study used high-end proteomic technology to examine the entire COL composition of the human IVD across fetal (developmental-FD), normal (healthy-ND), scoliotic (early degeneration-SD), herniated (degenerate-DH), and degenerated (DD) disk phenotypes.
Methods
Forty NP tissues were snap-frozen in liquid nitrogen (−196°C) immediately before being subjected to proteomic and bioinformatic analyses from five different disk phenotypes (eight each).
Results
Tandem mass spectrometric analysis revealed a total of 1,050 proteins in FDs, 1,809 in ND, 1,487 in SD, 1,859 in DH, and 1,538 in the DD group. Of 28 major collagens reported in the human body, this study identified 24 different collagens with 34 subtypes in NP. Fibril-forming collagens (COL-1, 2, and 11A1) and fibril-associated collagens with interrupted triple helices (COL-9A1, 12A1, and 14A1) were abundantly expressed in FDs, representing their role in the development of NP. Multiplexin (COL-15), a hybrid proteoglycan–collagen molecule, was discovered only in FDs. Degeneration was associated with COL2A1 downregulation and COL-10A1 upregulation.
Conclusions
COL10 was discovered to be a new biomarker for disk degeneration. Besides COL-1 and 2, other important COLs (6, 9, 11, 12, 14, 15) with anabolic potential and abundant expression in the fetal phenotype could be investigated for tissue engineering and novel DDD therapy.
Introduction
The largest aneural and avascular biological structure connecting vertebral bodies is the intervertebral disc (IVD). IVD pathologies are the most responsible for the global epidemic of chronic low back pain (LBP) [1,2]. Over the last 5 decades, there has been significant progress in understanding discal biology in health and disease, leading to the development of several preclinical and animal regenerative strategies to address LBP [3]. Collagen (COL), which accounts for about 25%–30% of the total protein content, serves as the template for any bone or cartilaginous tissue. IVD-related changes in COL composition have been linked to degenerative disc disease (DDD), and molecular targets are being investigated to restore the young, healthy COL phenotype [4,5]. However, despite the fact that vertebrates have 28 different types of COLs and over 40 collagen-like protein-coding genes, the majority of the IVD literature focuses solely on COL-1 and COL-2. This study aims to use high-end proteomic technology to analyze the entire COL composition of the human IVD across fetal (developmental phenotype), normal (healthy phenotype), scoliotic (early degeneration), herniated (degenerate phenotype), and degenerated (degenerate phenotype) IVDs to identify biomarkers for disc degeneration and molecular targets for novel therapy.
Materials and Methods
1. Collection of samples and processing
The research was conducted with the appropriate ethical clearance obtained from the institutional review board of Ganga Medical Centre and Hospitals, Coimbatore, India (IRB approval no., 2019/08/06). Nucleus pulposus (NP) tissues were isolated from IVDs from five different disc phenotypes. Under sterile conditions, fetal spines obtained from specimens following the medical termination of pregnancy were dissected for NP harvesting. This group (fetal disk phenotype [FD]) represented a proliferative and developmental phenotype with high proliferative potential. During an anterior release procedure, scoliotic disk phenotype (SD) was harvested from patients with idiopathic scoliosis. These disks represent an early degeneration phenotype because they are usually subjected to additional mechanical stresses while appearing normal on magnetic resonance imaging (MRI). MRI-normal IVDs obtained from brain-dead voluntary organ donors who had no history of LBP exhibited a healthy (normal) disk phenotype (ND). During surgical intervention, two types of samples were obtained for the degenerate group: one from herniated disk phenotypes (DHs) and the other from degenerated disk phenotypes (DD). Excised NP tissues were washed with phosphate buffer solution and immediately snap-frozen in liquid nitrogen (LN2; −196°C) before proteomic analysis. The demographic and Pfirrmann grades are enlisted in Table 1.
2. Extraction and digestion of proteins
Total proteins were extracted from cartilaginous NP tissue samples using buffers containing salts and detergents, as well as other protease inhibitors. Under aseptic conditions, 200 mg of tissue was pulverized and homogenized with LN2. RIPA (radio-immunoprecipitation assay) and 2% sodium-deoxycholate (SDS) buffers were used for the sequential extraction of hydrophilic and hydrophobic proteins, which were then quantified, cleaned up, and pre-fractionated on 10% SDS-PAGE (polyacrylamide gel electrophoresis), as described in our previous report [6].
3. Identification and analyses of proteins using Proteome Discoverer ver. 1.4
Tryptic digested peptides were purified and subjected to ESI-LC-MS/MS analysis at a concentration of 1,000 femtomole per injection performed in duplicate. The purified peptides were analyzed using an Orbitrap Velos Pro Hybrid mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) in positive ion mode electrospray ionization using an EASY-spray column (PepMap RSLC, C18, 2 μm, 100 Å, 75 μm×50 cm or 15 cm), as described earlier. Proteome Discoverer ver. 1.4 (Thermo Fisher Scientific) was used to identify proteins from raw data generated by tandem mass spectrometric analysis (.raw/.msf). By searching against the UniProt human database, the in-built SequestHT and Mascot search algorithms were used to match tandem mass spectra to peptide sequences. As reported previously [7], proteins with high peptide confidence and rank one were considered for identification after an effective screening of missed cleavages. The identified proteins were further checked for complementary DNA fragments, isoforms, uncharacterized entries, and normalized using the normalized spectral abundance factor method [8].
4. Collagen type analyses
By mapping the total proteins expressed in the study group (FD, ND, SD, DD, and DH) against the collagen list retrieved from the Matrisome database (http://matrisomeproject.mit.edu/other-resources/human-matrisome/) and other published literature [9–14], we were able to perform selective screening for all collagen types and their precursors. Protein data were post-processed by applying stringent parameters; collagen genes with peptide-spectrum match (PSM) ≥5 were selected based on their sample positivity restricted to 25%.
5. Functional enrichment analyses
To better understand the biological roles played by collagen types, ShinyGo (http://bioinformatics.sdstate.edu/go/) was used to analyze gene ontology data from selectively screened collagen (ver. 0.60). The functional pathway analysis was also carried out using the Reactome database pathway browser ver. 3.7 (https://reactome.org/). Cluster analysis between collagen and other components of the extracellular matrix (ECM) was conducted using Cytoscape ver. 3.8 (https://cytoscape.org/) with annotations derived from the Search Tool for the Retrieval of Interacting Genes ver. 11.0 (STRING; http://string-db.org) with default settings.
6. Validation by immunohistochemistry
Immunohistochemistry was used to validate the proteomic evidence of COL14A1. Before embedding dissected samples in paraffin, they were stored in formalin. The experiment used 2–5-m sections from paraffin blocks and the tree-step indirect method. Prior to rinsing with ethanol solution and blocking with 0.1% hydrogen peroxide, the blocks were subjected to antigen retrieval using Tris pH 9.5 and borate pH 8.0 in conjugation. A 1:500 dilution of COL14A1 polyclonal antibody (Thermo Fisher Scientific; #cat. no. PA5-49916) was used. Further sections were developed with DAB and Harris hematoxylin before being scanned at 400× magnification under a Leica DML light microscope (Leica Microsystems Ltd., Seoul, Korea) and the in-built Leica Application Suite ver. 4.5.0.418 software (Leica Microsystems Ltd.). The slides were graded as either negative or mild (+), moderate (++), or strong (+++) positive.
7. Statistical analyses
IBM SPSS software ver. 25.0 (IBM Corp., Armonk, NY, USA) was used to compare protein expression levels between sample groups. To understand the normal distribution of sample groups, a normality assessment was performed. In the case of a normal distribution, t-tests and one-way analysis of variance were used, and in the case of a violation of the normality assumption, Kruskal-Wallis/Mann-Whitney U tests were used. For the tests, the two-tailed alpha was set at 0.05.
Results
This study included 40 IVDs, eight in each group (FD, ND, SD, DD, and DH). Tandem mass spectrometric analyses revealed total proteins of 1,050 in FD, 1,809 in ND, 1,487 in SD, 1,859 in DH, and 1,538 in DD. Collagens, a major component of the ECM, were selectively screened in this study. Of the 28 major collagens reported in the human body, this study identified 24 major collagens with 34 subtypes (Table 2). After applying stringent cut-off filters (PSM ≥5, 25% sample positivity), 15 major types of collagens represented by 19 different collagen genes were analyzed further.

Comparison of various types of collagen reported in the literature and identified in our study population
1. Comparative analysis of collagen expressed across the study population (FD, ND, SD, DH, DD)
Major collagen expression varied across groups, with 12 collagens identified in the FD, eight in the ND, 14 in the SD, 16 in the DH, and 11 in the DD groups, respectively (Fig. 1A). A comparison of the study groups using http://bioinformatics.psb.ugent.be/webtools/Venn/ revealed that COL15A1 was specific to the FD group and COL19A1 was specific to the SD group. COL16A1 and COL3A1 were specific to the DH group. About eight different collagen genes (COL1A1, COL1A2, COL2A1, COL6A1, COL6A2, COL6A3, COL11A2, and COL14A1) were found to be common to all the groups (Fig. 1B).
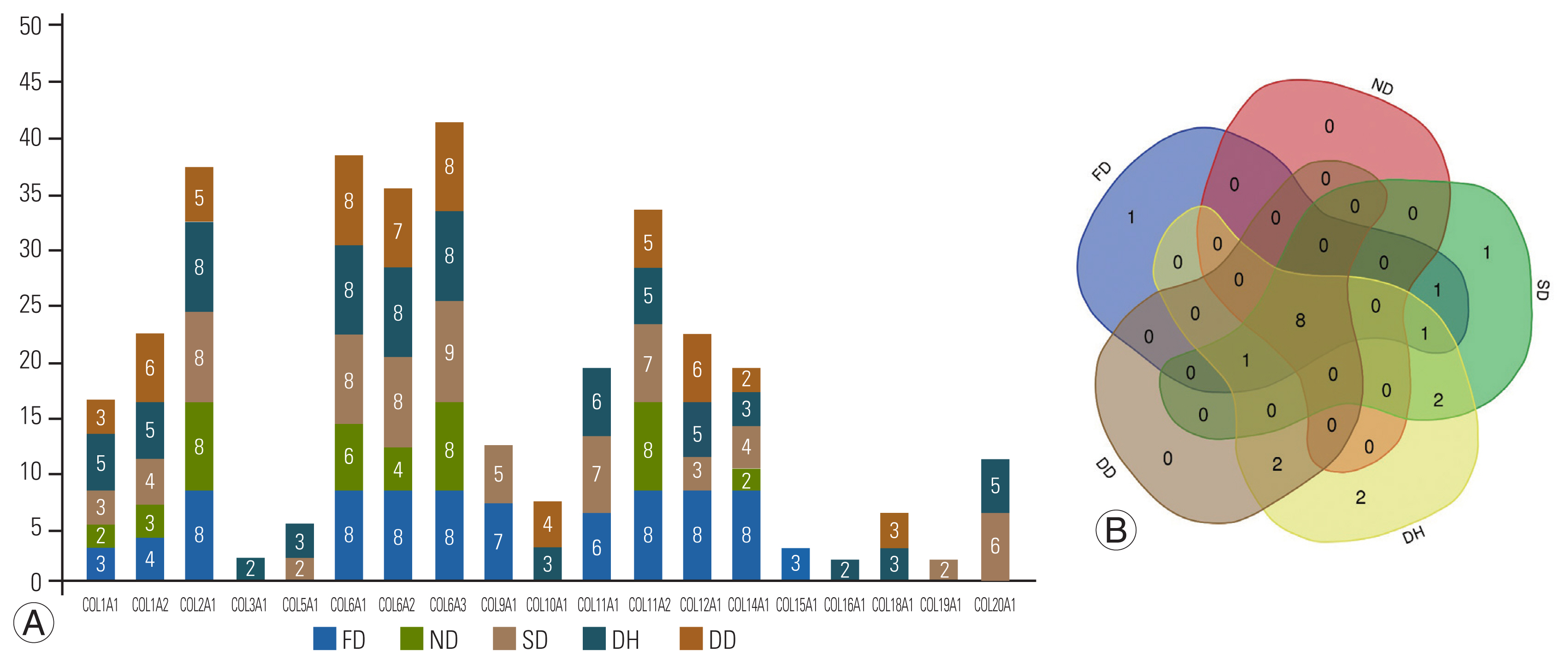
Collagen frequency detected in samples. (A) Stacked bar diagram represents frequency of detecting 19 identified different types of collagen protein in the study groups. The numbers within the stacked bars indicate the sample numbers (out of total eight discs evaluated under each group) expressing the specific collagen. (B) Venn diagram showing the overlapping collagen types between the groups (fetal [FD], normal [ND], scoliotic [SD], herniated [DH], and degenerated [DD] disk phenotypes) included in this study. About eight collagens were common to all groups. COL15A1 specific to FD group; COL19A1 specific to SD group; COL16A1and COL3A1 were found specific to DH group respectively.
2. Various types of collagens reported in our study
1) Fibril-forming collagens
The majority of the ECM is composed of fibril-forming collagens, and types 1, 2, 3, 5, and 11 were identified in our study (Fig. 2A–E). Collagen type-1 was the most abundant in fetal disks, with subtypes COL1A1 (FD versus ND, p-value=0.0281; FD versus DD, p-value=0.0005) and COL1A2 (FD versus DD, p-value=0.0019) being the most abundant. Collagen type-2 (COL2A1) showed the highest expression in FD, followed by SD and ND, with the lowest levels in diseased states. According to statistical analyses, COL2A1 levels were significantly higher in the FD group (p-value=0.0123) and the ND group (p=0.0043) than in the degenerate group. Collagen type-3 was discovered to be unique to the DH group, whereas collagen type-5 subtype COL5A1 was more common in the DH group. COL11A1 and COL11A2 were found in collagen type-11, with COL11A2 expression being higher in the FD group.
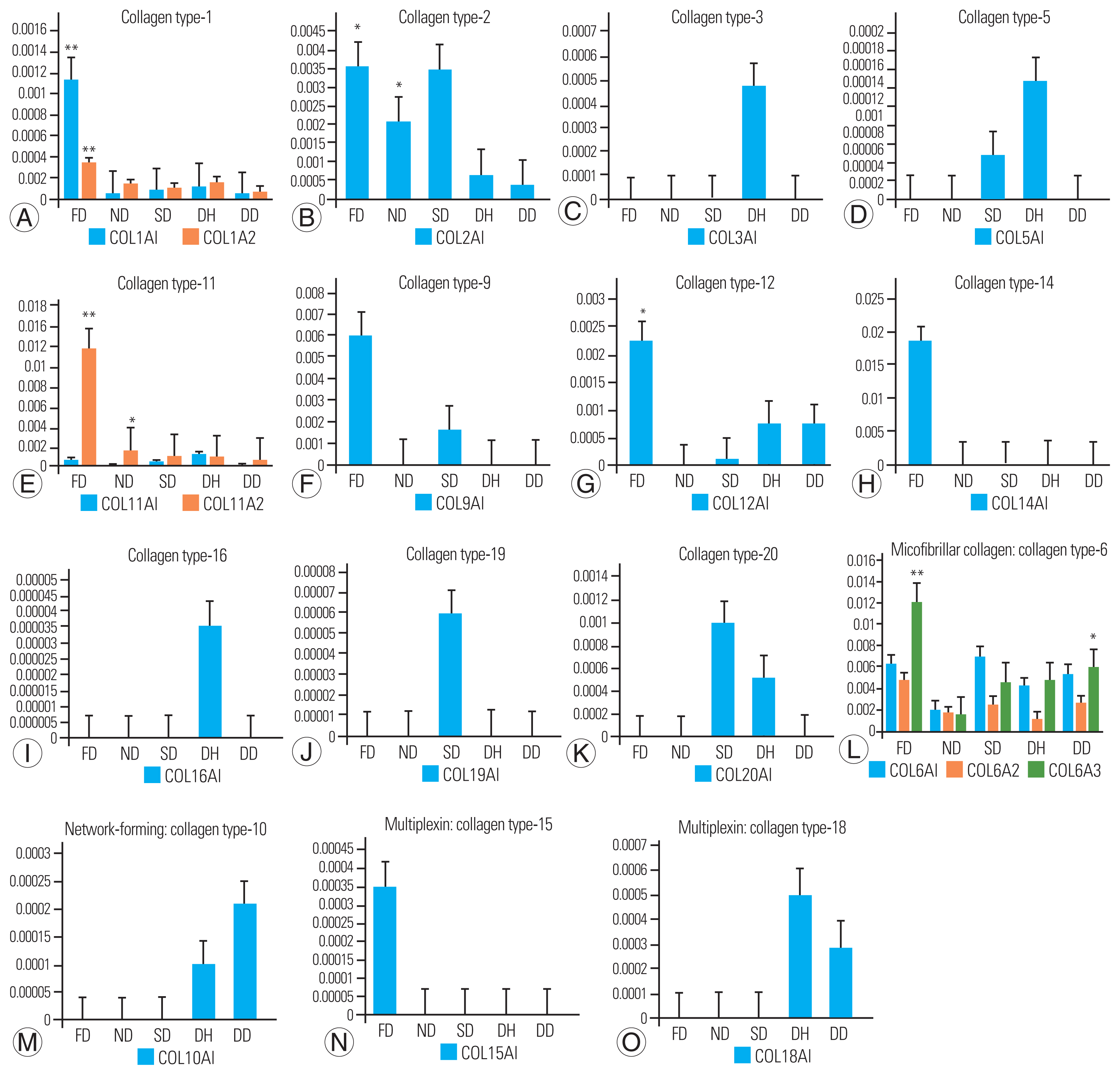
Bar graph representation of identified various collagen types across study groups. (A–E) Fibril-forming collagens: includes types such as -1, -2, -3, -5, and -11. Significant higher expression of COL1A1 was observed (fetal disk phenotype [FD] vs. normal disk phenotype [ND], p=0.0281; FD vs. degenerated disk phenotype [DD], p=0.0005) and COL1A2 in FD group (FD vs. DD, p=0.0019). COL2A1 was highly expressed in FD group with p-value, 0.0123 and with ND group (p=0.0043). Also, COL11A2 was found with increased expression in FD group against ND (p<0.0001) and DD (p=0.0002) groups, respectively. (F–K) FACIT collagen: includes types such as -9, -12, -14, -16, -19, and -20. Expression of COL9A1, COL12A1, and COL14A1 was found higher in FD group. Specific expression of COL16A1 and COL19A1 was found in herniated disk phenotype (DH) and scoliotic disk phenotype (SD) group. COL20A1 was found higher in SD followed by DH group. (L–O) Other types of collagens: includes microfibrillar/beaded filament collagen, network-forming collagen and multiplexins. COL6A1 was found higher in SD group; whereas COL6A2 and COL6A3 higher in FD comparatively. COL6A3 showed significantly higher expression against ND (p=0.0002). COL10A1 was higher in DD followed by DH. Multiplexin-COL15A1 was specific to FD whereas COL18A1 was higher in DD followed by DH. Y-axis represents average normalized spectral counts and the x-axis represents study group. (*) indicates statistical significance corrected using multiple statistical tests in IBM SPSS ver. 25.0 (IBM Corp., Armonk, NY, USA) with color codes (FD vs. ND, red; ND vs. DD, green, and FD vs. DD, violet).
2) Fibril-associated collagens with interrupted triple helices (FACITs)
The samples contained relatively short collagens with interrupted triple helices, as well as FACITs 9, 12, 14, 16, 19, and 20 (Fig. 2F–K). The FD group had higher levels of COL9A1, COL12A1, and COL14A1 expression. Although COL16A1 was found only in the DH group, COL19A1 was found only in the SD group. SD had the highest expression of COL20A1, followed by the DH group.
3) Other subtypes of collagen family
Microfibrillar/beaded filament collagen (collagen type 6: COL6A1, COL6A2, COL6A3), network-forming collagen (collagen type 10: COL10A1), and multiplexing collagen (COL15A1 and COL18A1) were also identified in our study (Fig. 2L–O). Although the COL6A1 subtype was abundant in SD, COL6A2 and COL6A3 were more prevalent in FD. DD had the highest level of COL10A1, followed by DH. Multiplexing revealed that COL15A1 was specific to FD, whereas COL18A1 was more prevalent in DD, followed by DH.
3. Functional enrichment analyses
ShinyGO was used to characterize the collagen types identified at the biological (Fig. 3A) and cellular levels to reveal their fundamental role (Fig. 3B). ECM organization and extracellular structure organization were found to be more significant in biological processes, with all identified collagen types (collagen types-1, 2, 3, 5, 6, 9, 10, 11, 12, 14, 15, and 18) participating. Other significant functions include collagen fibril organization along with various tissue morphogenetic processes. In the cellular component, collagens localized to the ECM, collagen-containing ECM, and extracellular space were found to be highly significant (p<0.05), supporting the nature of these collagens (Supplement 1).
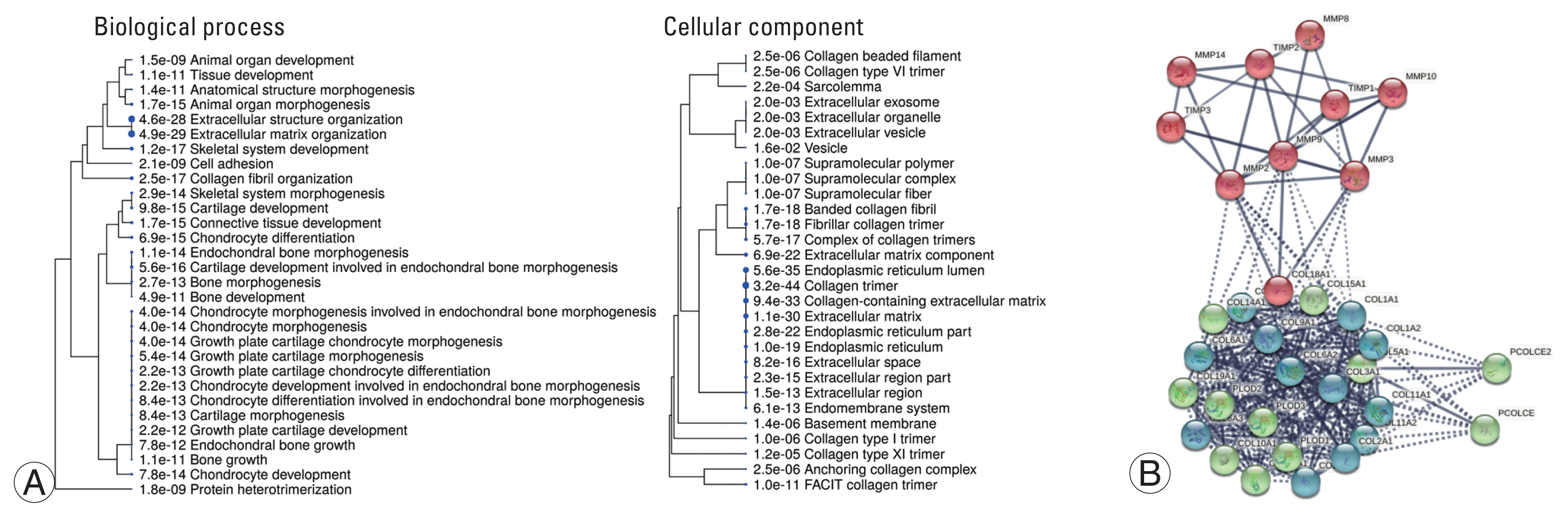
Functional analysis of collagens. (A) A hierarchical clustering tree demonstrates the correlation among biological process. Cellular component identified for observed collagen types along across study groups. Biological processes with many shared genes are clustered together. Bigger the size of dots indicates more significant p-value. In biological process, extracellular matrix organization and extracellular structure organization was found to be more significant along with other various tissue morphogenesis process. In cellular component, collagen was highly associated with endoplasmic reticulum-lumen and part, extracellular-matrix and space. Other components include basement membrane collagen trimer, fibril associated collagen with interrupted helices (FACIT) collagen trimer and collagen trimerization. (B) Protein-protein interaction network derived using STRING database (http://www.string-db.org) ver. 11.0 for observed collagens, collagenases (matrix metalloproteinases [MMPs]) and their inhibitors (tissue inhibitors of metalloproteinases [TIMPs]). The protein-protein interaction (PPI) network analysis showed a network of 49 nodes and 758 edges. The subnetworks were analyzed k-means clustering algorithm for three clusters had PPI enrichment p-value <1.0e-16 and average clustering coefficient 0.923. Nodes represent the input protein molecules whereas edges denote their inter-relationships imparting shared function.
An enriched pathway analysis using Reactome pathway browser ver. 3.7 (https://reactome.org/) yielded 96 pathways (Supplement 2). This study identified 33 significant pathways, as well as the proteins that participated in them (Table 3). When we mapped these identified collagen proteins in the pathway browser, we discovered 35 different collagen subtypes being mapped to various metabolic pathways, such as collagen formation, collagen biosynthesis and modifying enzymes, collagen chain trimerization, and collagen degradation, indicating their roles in various metabolisms. The interaction between identified collagens and matrix metalloproteinases (MMPs) was identified using the STRING database version 11.0 (https://string-db.org/), as shown in Fig. 3C. The association revealed three clusters: (1) the collagen cluster—all identified collagens in the study were found to be associated with procollagens (Procollagen C-Endopeptidase Enhancer [PCOLCE] and PCOLCE2); (2) the collagenase cluster—MMP-3, 8, 9, 10, and tissue inhibitors of metalloproteinases (TIMP)1; and (3) the inhibitor cluster—TIMP-2 and 3 were associated with MMP-2 and 14.
4. Quantitative abundance of procollagens: collagenases
Collagenases including MMP types 1, 2, 3, 8, 9, 10, 13, and 14, and cysteine proteases, such as cathepsin K, were identified. Although their basal expression results in normal collagen turnover, their excessive abundance can cause ECM degradation [15]. The abundance of procollagens to that of collagenases/metalloproteinases and their inhibitors is shown in Fig. 4. The expression of PCOLCE was found to be more in the FD group than in all other groups (p-value=0.011), followed by the expression of procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 (PLOD2). Six types of MMPs (2, 3, 8, 9, and 10) were found in the DD group, with high expression of MMP-8, followed by MMP-2, 9, and 10, which could cause excessive collagen degradation. MMP-14 was specific to the DH group along with commonly observed MMP-2 and 3. The ND group had MMP-3 and 10, where MMP-10 showed higher expression than all other groups; however, no statistical significance was observed. MMPs-3, 9, and 10 were found in the SD group. The presence of inhibitors (TIMP-1, 2, and 3) in the ND group was found to be comparatively higher than that in the DD group, which is essential to have a check on MMP expression. MMP expression can be triggered by various types of insults ranging from trauma and immune mechanisms to infection during adulthood. The FD group showed no evidence of MMP/TIMPs, indicating the absence of any insult triggering their expression.
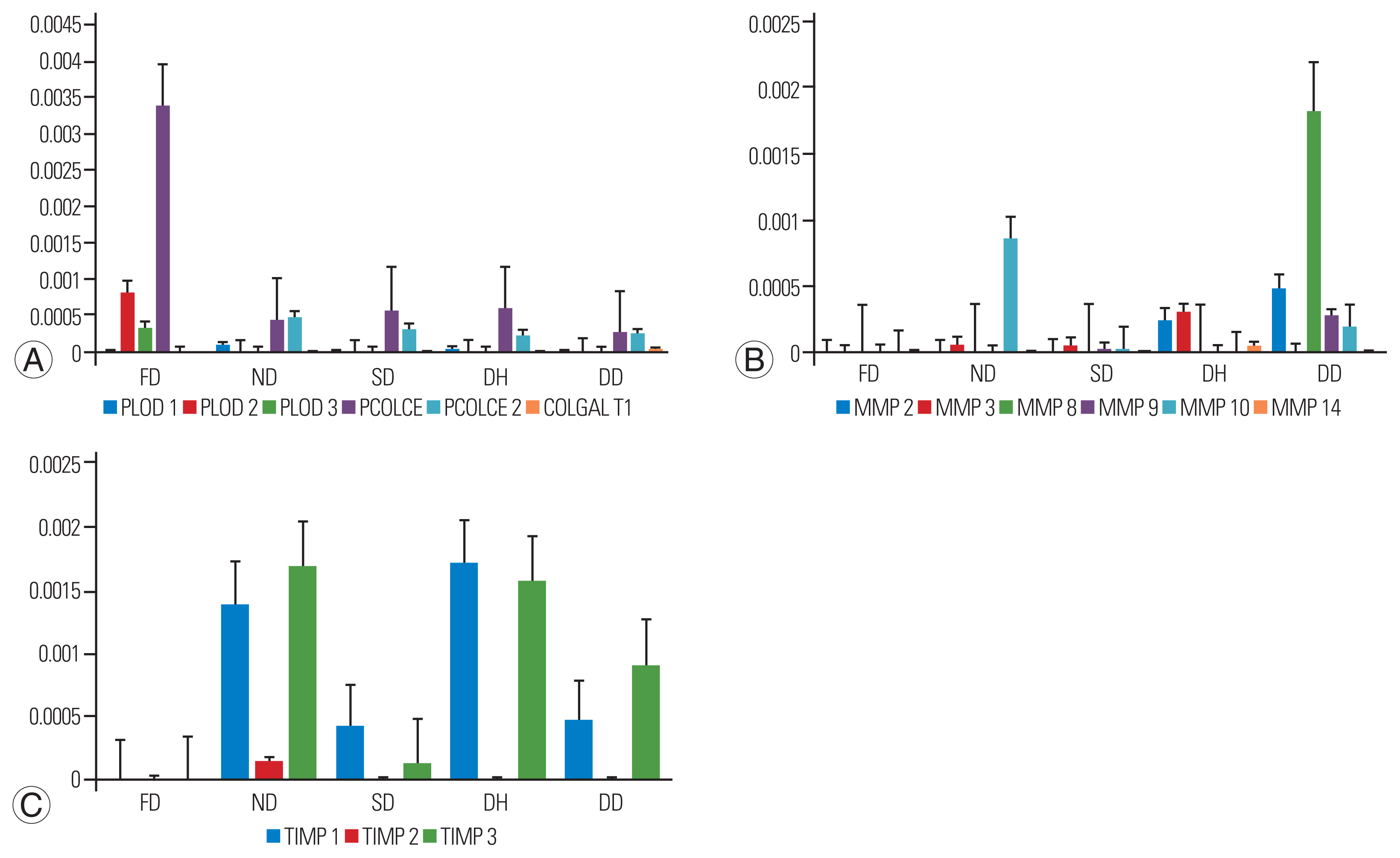
Bar plot representing the expression of procollagens, collagenases, and inhibitors across study groups (fetal [FD], normal [ND], scoliotic [SD], herniated [DH], and degenerated [DD] disk phenotype). (A) Procollagens: the expression of PCOLCE was found significantly higher in FD (with ND, p=0.021; DD, p=0.042) followed by the exclusive presence of PLOD2 and PLOD3. Lower expression of PCOLCE and PCOLCE2 were seen in other groups respectively. (B) Metalloproteinases/collagenases: DD group expressed all matrix metalloproteinases (MMPs) except MMP14. Interestingly, the expression of neutrophil collagenase: MMP8 was found higher compared to all other groups suggesting collagen degradation. Higher expression of MMP 10 was found in ND group, whereas lowest was observed in SD group; MMP 2 and 3 levels were slightly similar in DH group. (C) Inhibitors: tissue inhibitors of metalloproteinases (TIMP) 1, 2, 3 expression was observed in all the groups except FD group. However, the expression of inhibitors was found higher in ND when compared to DD group. Y-axis represents average normalized spectral counts and the x-axis represents study group.
5. Validation of COL14A1 expression
COL14A1, which was found to be abundant in fetal disks in this study, was also found to be abundant in bovine fetal disks in a previous study [16]. We chose immunohistochemistry to validate its expression as a suitable candidate for regenerative therapy using a new set of 10 fetal and five adult control disks. Seven of 10 fetal samples showed strong cytoplasmic and ECM staining. Adult samples, on the other hand, showed only minor staining in the ECM (Fig. 5).
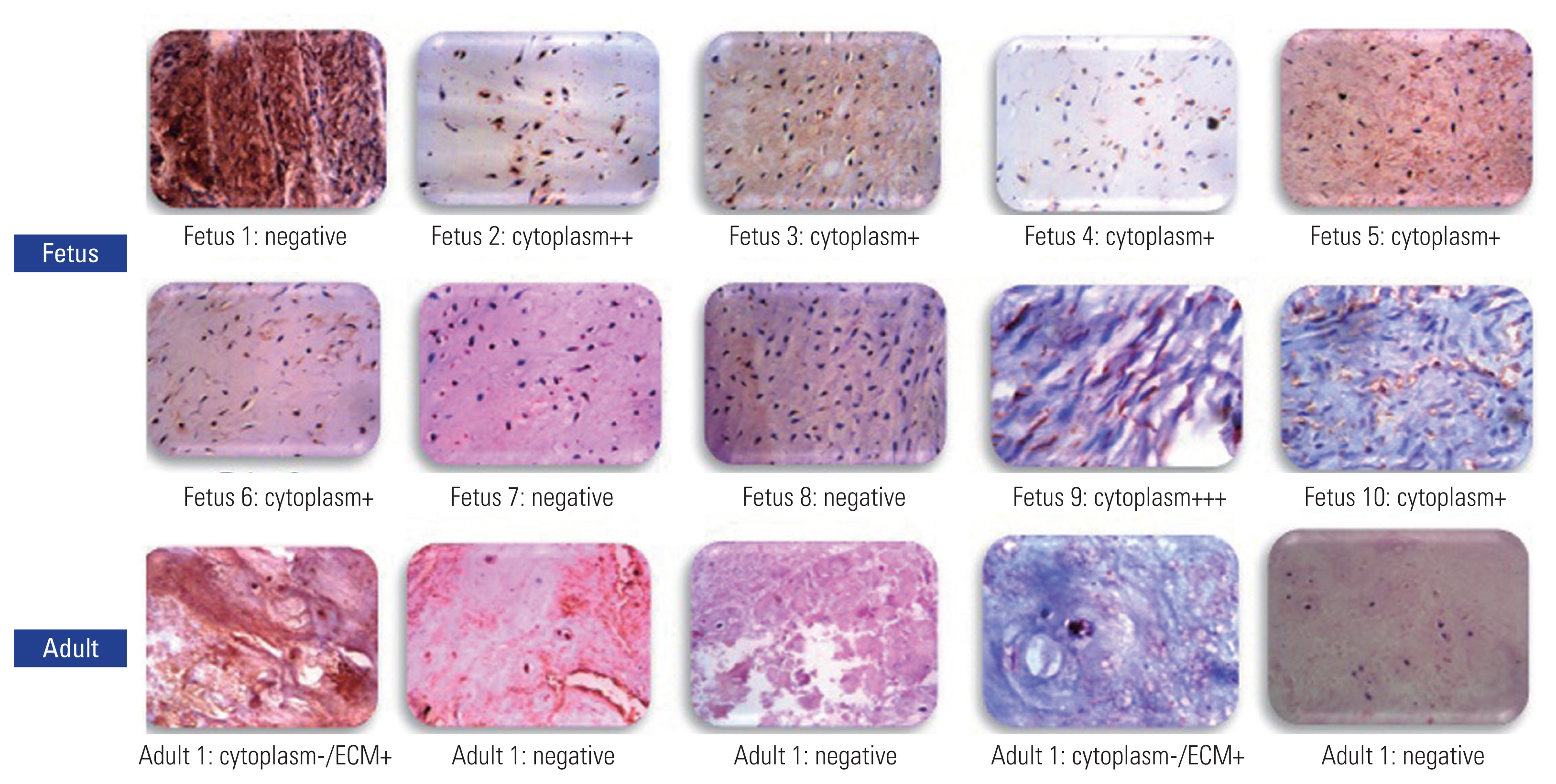
Validation of COL14A1 by immunohistochemistry. Ten fetal samples and five normal disk phenotype samples were used. Seven of fetal samples showed strong cytoplasmic and extracellular matrix (ECM) staining. In contrast, only two adult samples showed mild staining in ECM (magnification ×400, scale bar=50 μm). Positive samples are stained brown.
Discussion
Collagen degradation causes ECM disorganization and NP apoptosis, hastening the degenerative process [17]. The reversal of the COL-2 to COL-1 ratio in NP has been strongly linked to DDD [4]. Minimally invasive regenerative therapies are being investigated, including biological injections of COL-2 into IVDs, which have been shown to be effective in preventing and arresting DDD [5]. Collagen and its derivatives have been clearly found to have anti-inflammatory, antiapoptotic, and even regenerative effects in cartilage defects [18]. With current regenerative strategies focused at identifying the secretome of fetal and young healthy IVDs [19], we performed a selective proteomic analysis of collagen composition using a high-end mass spectrometer to unravel the complexity and diversity across five disk phenotypes representing embryonal, healthy, early, and late stages of disc degeneration.
We compared the collagen expression of fetal disks with that of healthy adult and degenerate disks to identify collagens with anabolic potential. We also compared healthy adult disks with degenerate disks to identify molecules essential for disk homeostasis and to establish biomarkers for disk degeneration. The precise biological roles of the differentially expressed collagens and the identified potential molecular targets are discussed comprehensively.
1. Collagen framework of young and healthy adult disks
Multiplexin (COL15A1), one of the most underreported collagens in humans, was the only collagen found to be specific to fetal disks, representing their role in IVD development. However, there is increasing evidence of its role in numerous physiological and pathological processes apart from maintaining the structural integrity of the ECM [20]. This collagen/proteoglycan hybrid molecule has chondroitin sulfate chains that holds water and is essential for the NP. It is interesting to note that zebrafish, which is one of the most studied organisms for its regenerative potential even in spinal cord injuries, has several paralogs of this COL15A1 and therefore, it seems to have definite anabolic potential [21].
Several studies have demonstrated the importance of COL-2 in maintaining healthy NP homeostasis [17]. We also observed a decreasing abundance of COL-2 with increasing age from fetus to scoliotic individuals, then healthy adults, and finally degenerate disks. COL-2 injections in ovine models have recently been discovered to have enormous regenerative potential [5]. With multiple randomized controlled trials documenting their role in chondral protection, COL-2 remains a definite molecular target for tissue engineering [22]. Although the reversal of the COL-1/2 ratio in NP is a definite biomarker of DDD, it is not known whether COL-1 is harmful to NP [23]. The daily consumption of COL-1 has been found to confer chondroprotective and anti-inflammatory roles and its highest expression in fetal NP in our study also supports these views and needs further research [18].
Another observation in the young developmental phenotypes was the higher expression of COL-6, 9, 11, 12, and 14. Similar to our study, Wu et al. [24] documented a higher abundance of COL-6, accounting for nearly 20% of NP collagen in fetal disks. Later, Roberts et al. [25] confirmed using immunolocalization that COL-6 is an important constituent of NP. With recent studies documenting its role in satellite cells, which proliferate and produce stem cells and aid in auto-regeneration of skeletal tissue, its role in IVD homeostasis needs to be studied in depth [26]. COL-9 is a well-documented adhesion protein in the ECM, where it becomes covalently cross-linked to COL-2 fibrils. Its deficiency in patients with diabetes and animal models have led to loss of ECM integrity and excessive degeneration and, therefore, is absolutely essential for disk homeostasis [27,28].
Caldeira et al. [16], in a study of bovine IVDs, compared fetal, young adult disks and aged phenotypes. They discovered that COL-12 and COL-14 were expressed only in fetal disks, which gradually disappeared in adulthood and observed COL-11 in young disks, which again decreased over age; therefore, they postulated that these molecules have regenerative potential [16]. In our study, though they were present throughout adulthood, their highest expression in fetal disks confirms earlier findings. As the difference in expression between fetal and adult and degenerate disks was the highest for COL-14 (present in only 25% of adult disks), we chose to validate this finding using immunohistochemistry and found intense staining of COL-14 in seven of 10 fetal disks, which were located more in the cytoplasm than in the ECM. Their presence in adults was only minimal and restricted to the ECM (Fig. 5).
2. Altered collagen signature in degenerate disks
When analyzing the degenerate disk phenotype, a striking observation was the presence of COL-18A1 only in the DH and DD groups. It has been reported as a normal constituent in the basement membrane of eyes [29]. However, elevated serum levels of endostatin, which is the cleavage product of COL-18A1, have been shown to reduce survival rates in individuals with pulmonary arterial hypertension [30] and its role in DDD is not known. Further, the upregulation of COL-10A1 and downregulation of COL-2A1 was associated with DDD. COL-10A1 has recently been designated as a novel biomarker of cartilage degradation and inflammation in patients with osteoarthritis [31]. Furthermore, its role in articular cartilage ossification could be compared to the conversion of healthy IVD to the fibrous degenerate type; thus, it could serve as a biomarker for DDD [32].
3. Strength of the study and limitations
Our study is unique in that we have documented the collagen expression of 40 IVDs across five different biological phenotypes in humans, which has never been done before. A comparative analysis between developmental, adult, and degenerate phenotypes has revealed novel biomarkers for degeneration and has also identified potential molecular targets for novel therapies in DDD. The proteomic finding was also validated using immunohistochemistry but was done only for COL-14. Our study has provided deep insights into the molecular understanding of IVD in health and disease.
4. Clinical significance
Our study used high-end proteomic technology for the first time to analyze the human IVD across five different biological states, revealing novel molecular targets for therapy and potentially changing the spectrum of care in LBP and DDD. LBP affects 80% of the population at least once in their lifetime, with a global age-adjusted point prevalence of 7.5% [33]. It is now considered a priority disease by the World Health Organization owing to increasing disability and loss of productivity. There is also a tremendous rise in the number of spine surgeries, resulting complications, and their associated costs. In the Netherlands, the average cost for a patient with chronic discogenic LBP is around EUR 7,911, but it is much higher in the United States [34]. Regenerative medicine has the potential to affect the entire spectrum of DDD. Improvements in the IVD structure have been demonstrated using cell culture models and animal models. However, given the enormous variability in biological mechanisms occurring at the molecular level, their translation into medical solutions has been limited.
Conclusions
The results of the study show the importance of collagens other than COL-1 and COL-2 in determining the health and disease of human IVDs. COL-6, 9, 11, 12, and 14 were identified to have higher expression in developmental phenotypes and are, therefore, of regenerative potential in addition to COL-15A1, which was uniquely present in fetal disks. Added to the downregulation of COL-2, the higher expressions of COL-18A1 and COL-10A1 were identified as novel biomarkers of DDD.
Supplementary Materials
Notes
This is the 2021 APSS-Asian Spine Journal Best Paper Award.
Conflict of Interest
No potential conflict of interest relevant to this article was reported.
Author Contributions
SR, DCR, CT, and MR conceived and formulated the project. SMN and CT contributed to the design of the analysis; performed lab experiments and bulk of data analysis; DCR and SMN wrote and prepared the manuscript. All authors have read through and given the final approval of the submitted publication.
Funding
The project was funded by Ganga Orthopaedic Research & Education Foundation (GOREF 2021-09).
Acknowledgements
We acknowledge the efforts of Ms M. Sujitha and Ms M. Dhanalakshmi for assistance in LC–MS/MS experiments.

