Genetic Markers for Idiopathic Scoliosis in Arab Population: A Pilot Study
Article information
Abstract
Study Design
Cross-sectional screening.
Purpose
This study was conducted to determine if there is any association of the three microsatellite markers on chromosome 19p 13.3 in unrelated Saudi Arabian girls who were suffering with adolescent idiopathic scoliosis (AIS) and their healthy siblings.
Overview of Literature
The genetic influence on the development of familial scoliosis has been previously described, but the genetic influence on AIS still remains unknown. Three microsatellite markers (D19S216, D19S894, and DS1034) of chromosome 19p 13.3 were reported to be significantly associated with familial scoliosis. This cross-sectional screening was carried out in AIS patients and their siblings.
Methods
For eleven Saudi Arabian girls who were treated for AIS and their 11 siblings, we performed a linkage analysis using parametric and nonparametric methods and using GENEHUNTER ver. 2.1. Multipoint linkage analysis was used to specify an autosomal dominant trait with a gene frequency of 0.01 at the genotypic and the allelic levels. One sided Fisher's exact tests were used in the analysis of the contingency tables for the D19S216, D19S894 and DS1034 markers.
Results
The analysis between the patient group and the healthy siblings showed that at the genotypic level there was a significant association of the markers and scoliosis (D19S894 [p=0.036], D19S216 [p=0.004], and DS1034 [p=0.013]). Yet at the allelic level, there was no statistically significant association of the markers between the AIS patients and their siblings.
Conclusions
Our pilot study shows that there is a genetic influence between the AIS patients and the siblings. We believe large scale genetic screening is warranted for the patients with AIS to identify beyond any doubt the influence of these markers.
Introduction
The most common deformity in a growing spine is scoliosis and girls are affected more than the boys1. The incidence of scoliosis in Saudi Arabia was reported to be in the range of 0.16-0.5%2,3 and 59% of these cases were adolescent idiopathic scoliosis (AIS)4. McCarthy5 suggested that the long term cardio-pulmonary complications due to scoliosis can be prevented if an early diagnosis is made and the progression of the curves can be prevented. In our culture, girls remain covered and their deformities often go unnoticed; hence, they present late and surgery remains the only option with a high rate of complications6.
The genetic influence in familial scoliosis has been reported by a number of researchers7-9, but Chan et al.10 scanned >400 fluorescent microsatellite markers and found three markers (D19S216, D19S894, and D19S1034) linked to chromosome 19p 13.3 in the children with familial scoliosis and this was confirmed by Alden and their colleagues11. It was reported that the genetic differences between individuals from different human populations closely resemble each other and these genetic differences do not differ more than those between unrelated subjects within a single population12,13. We postulated that a pilot study could be limited to only three microsatellite markers on chromosome 19p 13.3. With this objective, we conducted this study on the three microsatellite markers on chromosome 19p 13.3 in 11 unrelated Saudi Arab girls who had AIS and we compared their findings with those of their siblings.
Materials and Methods
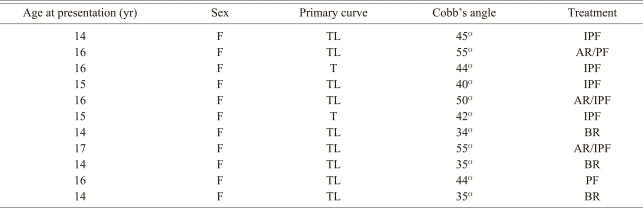
We studied 11 consecutive unrelated ethnic Saudi Arabian girls living in the Eastern Province of Saudi Arabia and all the girls had AIS and a Cobb angle of ≥30 degrees (Table 1). The 11 girls had routine radiographs and bending films, magnetic resonance imaging of the spine and pulmonary function tests. A comparative group of 11 siblings who were not affected with any spinal disease were classified as "phenotype unknown" for the linkage analysis. As all the patients and adults were ≤18 years of age, informed consent was obtained from the parents.
Genomic DNA was extracted from the peripheral blood and the microstattelite markers D19S216, D19S894 and D19S1034 were studied to determine if there was any significant association of the three microsatellite markers of chromosome 19 between the patients and their siblings. The fluorescent amplification products were then analyzed by capillary electrophoresis using a genetic analyzer 3130 from Applied Biosystems to determine the size of the amplified product by making comparisons with a size marker. Linkage analyses were performed, using parametric and nonparametric methods, by the use of GENEHUNTER ver. 2.1 (Kruglyak et al.14). The allelic frequency of each marker was calculated from the database. Two-point linkage analysis was performed with the MLINK software of the LINKAGE package ver. 5.215. The microsatellite marker locations were specified according to databases of the Center for Medical Genetics, the Marshfield comprehensive human linkage map, the National Center for Biotechnology Information, the Whitehead Institute/MIT Center for Genome Research, the Genome Database and the Stanford Human Genome Center Web sites. Multipoint linkage analysis was used for specifying an autosomal dominant trait with a gene frequency of 0.01 and an estimated penetrance of 80%. These parameters were taken from Chan et al10. One sided Fisher's exact tests were used in the analysis of the contingency tables for the D19S216, D19S894, and D19S1034 markers. A p-value <0.05 was considered as statistically significant. This study was approved by the research committee of the College of Medicine, King Faisal University, Dammam and King Fahd University Hospital, Alkhobar.
Results
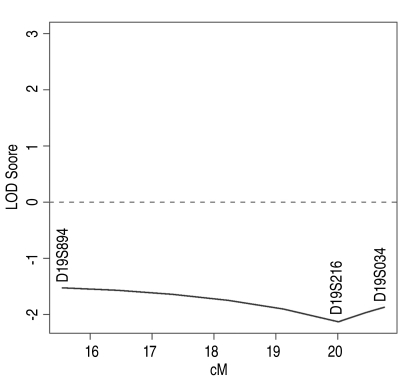
Twenty-two girls with 11 with AIS and 11 siblings were studied. The parents were unrelated and there was no history of AIS in any of the families. The three microsatellite markers with their allelic IDs and frequencies were studied on chromosome 19p 13.3. Tables 2-4 gives the analysis at the genotypic level. It showed that at the genotypic level there was significant association between the markers and scoliosis (D19S894 [p=0.036], D19S216 [p=0.004] and DS1034 [p=0.013]). Yet at the allele level, there was no statistically significant association of the markers between the AIS patients and their siblings (D19S894 [p=0.155], D19S216 [p=0.07] and DS1034 [p=0.34]) (Tables 5-7). As shown in Fig. 1, we did not find any significant linkage between the markers and scoliosis using a model-based approach. Indeed, all the non parametric LOD scores were equal to 0.

The contingency table below shows the numbers of the marker D19S894 that occurred at the genotypic level

The contingency table below shows the numbers of the marker D19S1034 that occurred at the genotypic level

The contingency table below shows the numbers of the marker D19S894 that occurred at the allelic level
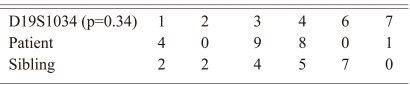
The contingency table below shows the numbers of the marker D19S1034 that occurred at the allelic level
Discussion
This pilot study involved 22 Arab girls and it showed that for patients with AIS the genetic markers can be targeted for making an early diagnosis of scoliosis and so this can prevent extensive surgery in the majority of AIS sufferers. We found that at the genetic level, the presence of these markers in AIS patients was statistically significant when compared to that of the siblings. All three markers (D19S216, D19S894, and D19S1034) were present on chromosome 19p 13.3 among all the ethnic Saudi Arabian girls. Our findings are not similar to the report of Chan et al.10 in their study of the families with adolescent idiopathic scoliosis and there was significant differences both at the genetic and the allele levels in that study, whereas in our small study, there was significance only at the genetic level. Miller et al.9 suggested that the area of future study for familial AIS is within the region of chromosomes 6,9,16, and 17. Raggio et al.16 recently found evidence of an AIS susceptibility locus on chromosome 12.
The mode of inheritance of AIS is still not clear even after extensive study of the human genome. Emery and Remoin17 believed that the genetic influence in AIS follows the principles of the Mendelian Inheritance, but Lowe et al.18 suggested that there is evidence that AIS is inherited in an autosomal dominant manner. The study by Justice et al.8 sheds light on the aspect that there is a possibility of X-linked inheritance in AIS. Scoliosis has long been know to occur in the human race and scientists have yet to determine the exact mode of inheritance.
The majority of the genetic studies on AIS has been reported series on families and twins19 with one type of genetic influence superseding the other genes. Although there is considerable evidence in the literature with regard to the familial relationships and the probability of developing AIS or having AIS, this probability is relatively low in many cases. According to our institutional survey of all the treated AIS patients, ≤1% had a history of AIS in another sibling. Adolescent idiopathic scoliosis occurs in patients with no family history as well, and the incidence of familial scoliosis is low for the majority of our ethnic Arab population. To find the genetic influence, we compared the genetic markers of the sufferers with those genetic markers of their siblings. It would be ideal if we could identify genetic markers that could be used among the general population to predict the development of AIS.
The limitations of this study are the study group was small and since we had no cases of familial scoliosis, the comparison had to be made with the siblings. Additionally, this study was done in one region of the country and there could be differences in the significance of the markers from other regions as well. In conclusion, we believe that in the patients with non-familial AIS, the genetic locus is linked to chromosome 19p 13.3 and further studies with larger numbers of patients are needed to confirm this locus in AIS patients.